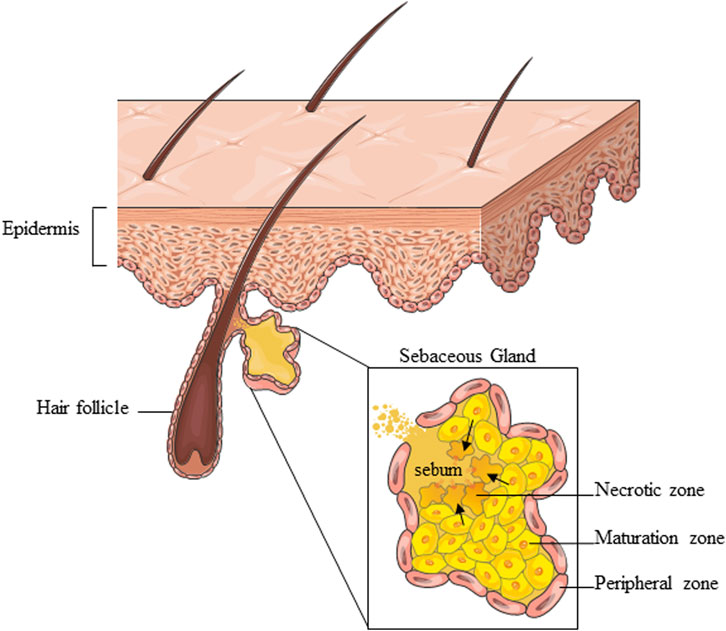
Sebaceous glands (SG) and their oily secretion, sebum, are essential for maintaining skin structure and function. Deregulation of SGs can lead to skin disorders such as acne and may also impact immune responses and energy metabolism.
This study utilizes sebaceous hyperplasia and integrates spatial transcriptomics, pseudotime analysis, RNA velocity, and functional enrichment to map the landscape of sebaceous differentiation, offering the first high-resolution spatial portrait of the SG transcriptional landscape.
Methods
This study used spatial transcriptomics (ST) to analyze sebaceous hyperplasia, characterized by increased size and number of glandular acini without structural abnormalities. Sequencing data were visualized using Uniform Manifold Approximation and Projection (UMAP), and unsupervised clustering identified different cell types. Self-Organizing Maps (SOMs) and gene set (GS) analysis were employed to characterize gene expression. Pseudotime and RNA velocity analyses mapped the progression of sebocyte differentiation. Validation was performed by comparing the results with published SG transcriptome data and protein expression patterns from the Human Protein Atlas.
Key Detailed Findings
Identification of Sebocyte Differentiation Stages: Unsupervised clustering identified four sebocyte differentiation stages (SEB-B, SEB-1, SEB-2, SEB-3) and six non-sebocyte cell types. The differentiation pathway was visualized using UMAP, showing a progression from SEB-B (proliferation) to SEB-3 (cell death).
Gene Expression Modulation: Transcriptome analysis revealed specific gene modules associated with each sebocyte stage. SEB-B showed high expression of cell cycle and oxidative phosphorylation genes, SEB-1 and SEB-2 were involved in lipid metabolism, and SEB-3 was associated with apoptosis-related functions.
Pseudotime and RNA Velocity: Pseudotime analysis confirmed a continuous differentiation trajectory from SEB-B to SEB-3, with RNA velocity vectors indicating a loss of transcriptional activity as sebocytes matured. This progression was corroborated by changes in gene expression patterns along the trajectory.
Validation with Independent Data: Protein expression patterns from the Human Protein Atlas confirmed the spatial localization of sebocyte-specific transcripts. Comparison with recent SG transcriptomics studies showed significant overlap in differentially expressed genes, supporting the identified sebocyte development stages.
Functional Implications: The study identified novel candidate genes involved in sebocyte differentiation, providing potential targets for modulating SG function in skin diseases. The findings offer a comprehensive understanding of sebaceous differentiation and suggest new experimental approaches for translational research.
This study provides a detailed characterization of the transcriptional dynamics in sebaceous gland differentiation, validated by independent data sources, and identifies potential molecular targets for therapeutic intervention in SG-related skin disorders.
Link to Article: https://tinyurl.com/mpmwa6fh