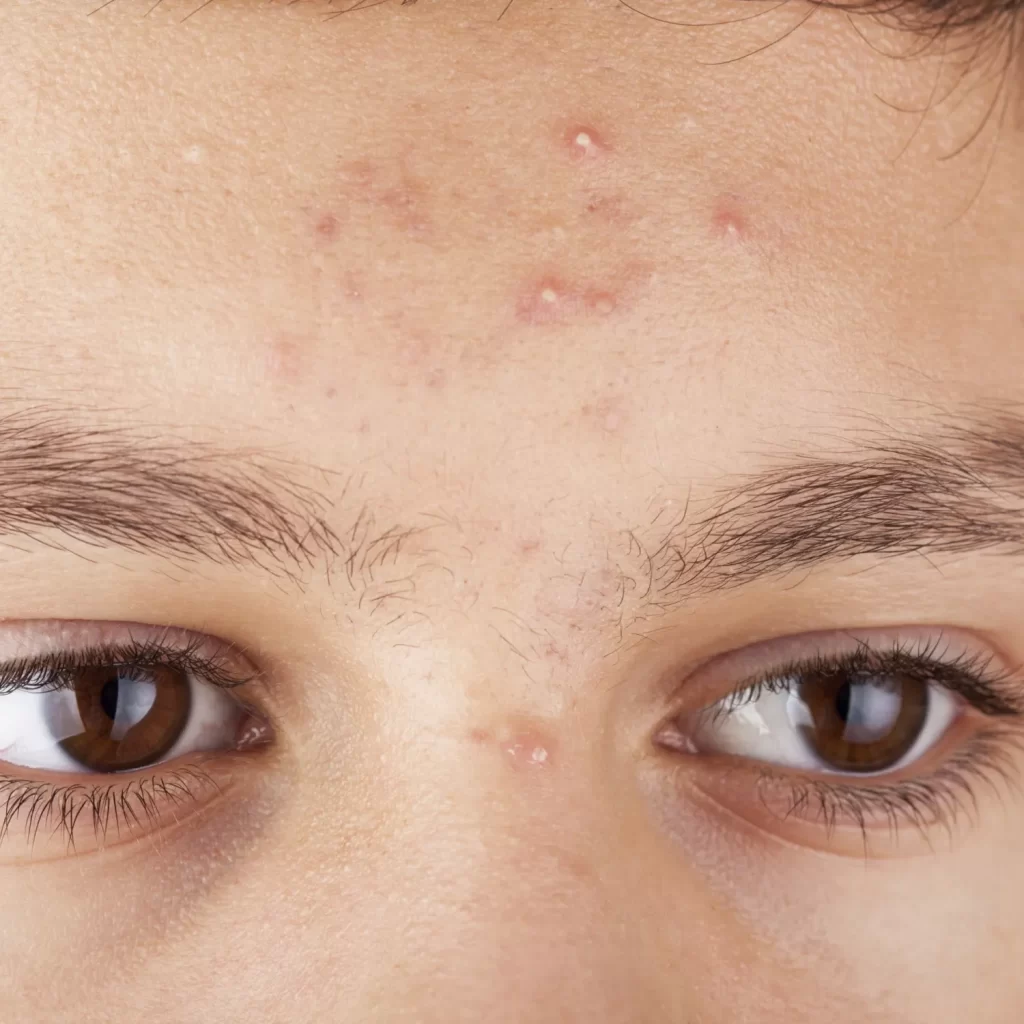
Acne vulgaris is a highly prevalent dermatological disorder, affecting over 85% of adolescents and often persisting into adulthood, particularly among women. Although not typically life-threatening, untreated acne carries significant physical and psychological consequences. Notably, acne is the only dermatological disorder identified as a risk factor for suicide, especially among men. The complexity of acne pathogenesis, which involves a multi-factorial interplay of genetic, hormonal, environmental, and skin microbiome factors, remains incompletely understood. This lack of comprehensive understanding contributes to variability in clinical presentation and response to treatment.
To overcome the limitations of traditional research, the complex and multifactorial nature of acne necessitates a multi-omics approach, which single-layer analysis cannot provide. Omics technologies—such as genomics, transcriptomics, proteomics, and metabolomics—represent large-scale biological analyses that provide new opportunities to understand acne pathogenesis. By enabling researchers to study the complex interactions among genes, proteins, and metabolites that drive the disease, these approaches support the development of targeted therapies. The use of multi-omics provides highly detailed insights into an individual’s biological characteristics, thus facilitating the realization of precision medicine—an approach that individualizes diagnosis and treatment based on a patient’s molecular profile.
Key Findings
The comprehensive review demonstrated that large-scale molecular analysis offers a deeper understanding of the mechanisms underlying acne vulgaris. Key findings across the different omics disciplines include:
• Genomics:
◦ A genome-wide meta-analysis identified 29 new genetic loci linked to acne susceptibility, bringing the total known risk loci to 46. These genes are associated with inflammation, cell adhesion, and hair follicle structure.
◦ Genetic factors based on these loci contribute approximately 5.6% to an individual’s risk of developing acne.
◦ Differences between skin-colonizing and infection-associated Cutibacterium acnes (C. acnes) strains are rooted in genes involved in metabolism and DNA repair, rather than classical virulence factors.
◦ Specific genetic variants in the TNF-α promoter region were associated with increased TNF-α expression and elevated inflammation, correlating with increased insulin resistance in patients with severe acne.
• Transcriptomics:
◦ Inflammatory biomarkers such as IL-1β, CXCL8, and SPP1 emerged consistently as crucial elements linked to acne development and promising therapeutic targets.
◦ Transcriptomic analysis of acne-prone, non-lesional skin indicated early dysregulation in innate immunity and epidermal barrier function.
◦ Pathway analysis suggested enhanced metabolic activity in non-lesional skin and identified mTOR inhibitors as potential agents to modulate gene expression.
◦ Single-cell data revealed increased basal cells and macrophages in acne lesions, and highlighted gender differences influencing cellular abundance and immune signaling.
• Proteomics:
◦ The Propionibacterium acnes phylotype associated with acne induces significantly higher levels of IFN-γ and IL-17 (up to three times greater) compared to phylotypes from healthy skin, and expresses higher adhesion proteins.
◦ Antibiotic-resistant C. acnes strains showed elevated levels of lipases and the cell division protein FtsZ, suggesting FtsZ as a potential therapeutic target.
◦ By integrating plasma proteome and GWAS data, novel protein biomarkers were identified, including FSTL1 and ANXA5, which showed strong causal associations and increased acne risk.
• Metabolomics:
◦ Acne patients show significant metabolic profile differences, including lower levels of essential and non-essential amino acids compared to healthy controls.
◦ Gender influences metabolic pathways; female acne patients exhibited higher levels of DHEA-S and free fatty acids (FFA) than males.
◦ Sebum analysis showed significant lipid alterations in adolescents with acne, including increased levels of triglycerides, squalene, and MUFAs, with squalene levels correlating with comedone count.
The future implications of these findings are critical for advancing precision medicine in dermatology. By providing detailed molecular insights into individual variability—such as immune dysregulation, sex-specific immune responses, and functional single-nucleotide polymorphisms—multi-omics enables the development of personalized treatment strategies tailored to an individual’s genetic profile, disease severity, and ethnic background. Multi-omics data facilitates the identification of novel biomarkers and provides better insight into pathological pathways. However, further studies are needed to validate these molecular findings and translate them into improved strategies for the diagnosis, prevention, and treatment of acne vulgaris. Continued integration, supported by emerging technologies like cloud computing and big data analytics, is essential to ensure that complex data leads to improved clinical outcomes.
Link to the study: https://tinyurl.com/mspx5sa3